Introduction: Bridging Massive Data to Precise Insights
The biomedical field is generating vast amounts of data at an unprecedented rate – from gene sequences and protein structures to drug responses and clinical trial results. This data holds crucial clues to unraveling the mysteries of life and conquering diseases. However, effectively extracting valuable knowledge from this complex, heterogeneous data and applying it to clinical decision-making and drug development remains a significant challenge.
Large Language Models (LLMs) have demonstrated powerful capabilities in processing and understanding textual information. Still, their knowledge is primarily derived from training data, which can sometimes lead to inaccuracies or “hallucinations” inconsistent with established scientific knowledge. To address this, BIT Research Alliance is actively exploring the integration of Knowledge Graphs (KG) with Retrieval Augmented Generation (RAG) techniques, forming the KG-RAG framework. This aims to provide LLMs with more reliable and structured knowledge support, especially in fields requiring high precision, such as bioinformatics and personalized medicine.
Knowledge Graphs (KG): The Cornerstone of Structured Biomedical Knowledge
A Knowledge Graph is a way of representing knowledge in a graphical structure, composed of “Nodes” and “Edges”:
- Nodes: Represent real-world entities, such as:
- Diseases (e.g., Acute Monocytic Leukemia, Osteosarcoma)
- Genes (e.g., RUNX1, SPRED1, ELANE)
- Drugs
- Proteins
- Symptoms
- Treatment methods
- Edges: Represent various “relationships” between entities, for example:
- [Gene A] is associated with [Disease B]
- [Drug X] is used to treat [Disease Y]
- [Protein P] is encoded by [Gene G]
In this manner, a knowledge graph can organize scattered biomedical information into an interconnected network of knowledge, clearly revealing the complex relationships between entities.
Large Language Models (LLM) and Retrieval Augmented Generation (RAG)
- Large Language Models (LLM): As previously mentioned, LLMs are powerful in natural language processing, but their knowledge updates and accuracy are limited by their training data.
- Retrieval Augmented Generation (RAG): RAG is a technique that allows an LLM to retrieve relevant information from an external knowledge base before generating a response. This enables the LLM’s answers to be based on more current and specialized knowledge, rather than solely on what it learned internally.
KG-RAG: A Powerful Combination of Knowledge Graphs Empowering LLMs
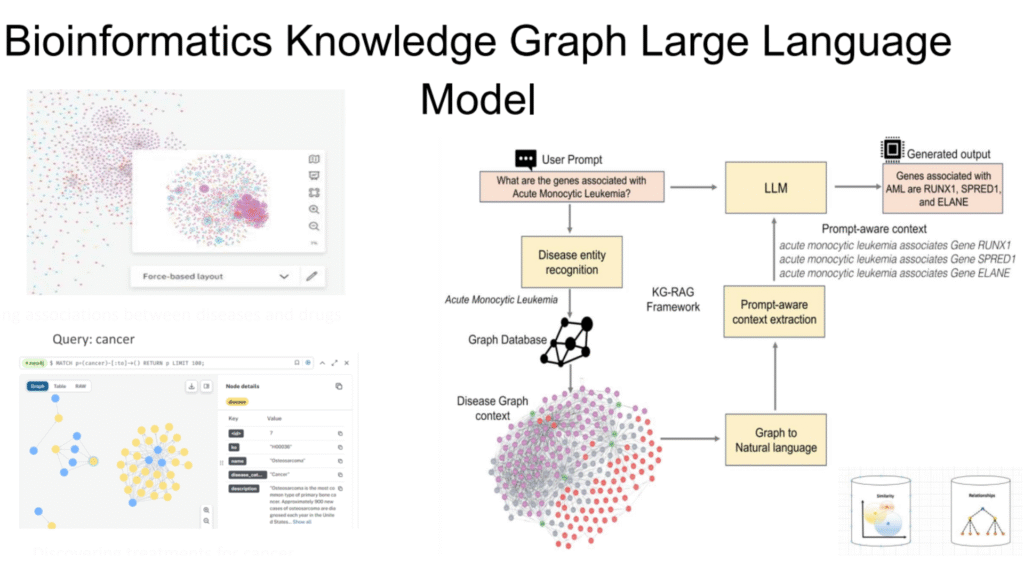
The core idea of the KG-RAG framework is to use the knowledge graph as the external knowledge base for the LLM in the RAG technique. The operational flow is roughly as follows :
- User Prompt: For example, “What genes are associated with Acute Monocytic Leukemia?”
- Disease Entity Recognition: The system first identifies key medical entities from the prompt, such as “Acute Monocytic Leukemia.”
- Knowledge Graph Retrieval (KG-RAG Framework – Prompt-aware context extraction):
- Utilizes the identified entity to query the knowledge graph database (Graph Database).
- Retrieves nodes and edges related to this entity, forming a “subgraph” or “context” highly relevant to the prompt (Disease Graph context). For instance, finding all gene nodes associated with “Acute Monocytic Leukemia.”
- Context Injection and LLM Generation (LLM + Prompt-aware context):
- Injects the relevant contextual information retrieved from the knowledge graph (e.g., “Acute Monocytic Leukemia is associated with Gene RUNX1,” “Acute Monocytic Leukemia is associated with Gene SPRED1”) into the prompt given to the LLM.
- The LLM generates the final answer based on the original prompt and the injected knowledge graph context.
- Generated Output: For example, “Genes associated with Acute Monocytic Leukemia are RUNX1, SPRED1, and ELANE.”
BIT Research Alliance’s KG-RAG Applications in Bioinformatics and Personalized Medicine
At BIT Research Alliance, we are actively applying the KG-RAG framework to the following key areas:
- Precise Analysis of Disease-Gene/Drug Associations :
- Quickly and accurately answering questions about the associations between specific diseases and genes, drugs, or other biomolecules.
- Discovering potential drug targets or new disease biomarkers.
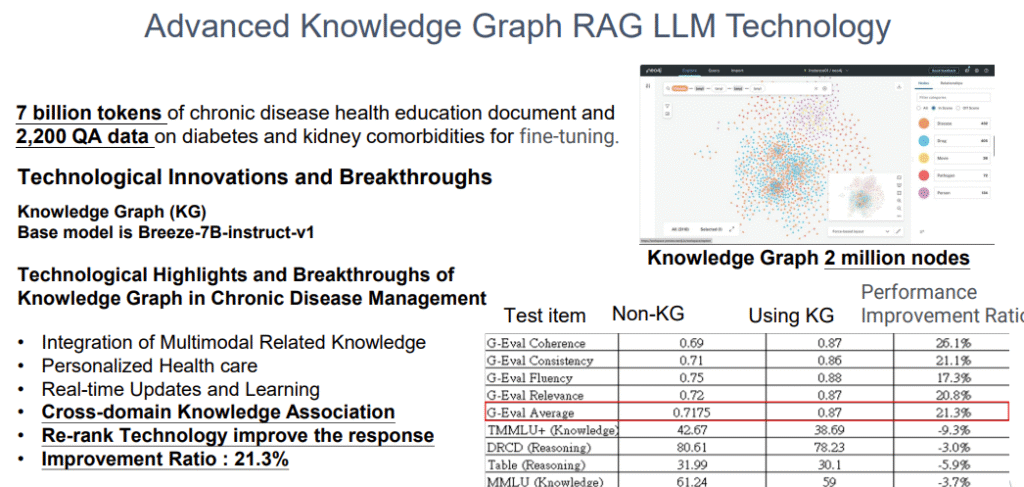
- We utilize a knowledge graph with millions of nodes , combined with a fine-tuned LLM (like Breeze-7B-instruct-v1), to significantly improve the accuracy of related knowledge Q&A. The G-Eval average score increased from 0.7175 (without KG) to 0.87 (with KG), an improvement of 21.3%.
- Personalized Treatment Regimen Recommendations:
- Recommending safer and more effective personalized treatment plans by combining a patient’s genotype, medical history, and information from the knowledge graph regarding drug responses and side effects.
- Accelerating Drug Discovery and Development:
- Assisting researchers in discovering new drug mechanisms of action or candidate drugs by analyzing complex relationships between compounds, protein targets, and diseases within the knowledge graph.
- Enhancing Medical Literature Understanding and Knowledge Discovery:
- Helping researchers quickly extract key entities and their relationships from massive volumes of medical literature, thereby building and expanding medical knowledge graphs and uncovering new scientific insights.
Advantages and Value of KG-RAG
- Improved Accuracy and Reliability: Introducing structured, validated knowledge graphs effectively reduces the likelihood of LLMs generating “hallucinations,” making their answers more scientifically sound.
- Enhanced Interpretability: By tracing the knowledge graph paths that an LLM’s answer is based on, its reasoning process can be better understood, increasing model trustworthiness.
- Continuous Knowledge Updates and Expansion: Knowledge graphs can be updated and expanded independently of the LLM, ensuring that the LLM has access to the latest medical knowledge.
- Handling Complex Queries: Better able to handle queries that require multi-step reasoning or involve complex relationships between multiple entities.
Challenges and Future Outlook
Despite its immense potential, the development of KG-RAG still faces challenges, including the cost of constructing and maintaining high-quality medical knowledge graphs, efficient fusion techniques between KGs and LLMs, and handling potential noise and inconsistencies within KGs.
BIT Research Alliance will continue to invest in R&D to optimize knowledge graph construction methods, enhance the efficiency and robustness of the KG-RAG framework, and explore its applications in a broader range of biomedical fields. We believe KG-RAG will become a crucial bridge connecting massive biomedical data with precise clinical insights, making significant contributions to achieving true personalized medicine and accelerating medical breakthroughs.
Conclusion: Knowledge Empowers Intelligence – KG-RAG Leads a New Chapter in Precision Medicine
The combination of knowledge graphs and large language models, particularly through the KG-RAG framework, presents unprecedented opportunities for bioinformatics and personalized medicine. BIT Research Alliance is committed to leveraging this powerful technological combination to extract deep knowledge from complex biomedical data and translate it into practical applications that can improve patient quality of life and drive medical progress. We look forward to collaborating with partners from all sectors to jointly usher in a new era of precision medicine empowered by knowledge.
